|
Labeling of DNA probes for in situ hybridization is commonly performed by PCR-incorporation of labeled dUTP or dCTP or by an enzyme mixture of DNase I / DNA polymerase I (termed Nick Translation) (Tab. 1, Morrison et al. 2003, Wiegant et al. 2001).  More information... More information...
Check out our enzymatically incorporable Fluorescent, Hapten- , Amine- or CLICK- modified dUTPs or dCTPs as well as corresponding labeling kits!
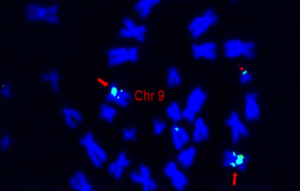
Figure 1: Chromosome 9 is visualized by fluorescence in situ hybridization (FISH). The ATTO-488-labeled DNA probe was generated from BAC clone BAC268E1 by nick translation with Aminoallyl-dUTP-XX-ATTO-488.

Jena Bioscience is a primary manufacturer offering
- both labeled single nucleotides and kits
- bulk amounts
- custom formulations, packaging and labeling
 Download our new DNA labeling brochure Download our new DNA labeling brochure
Table 1: Selected product references. BAC: bacterial artificial chromosome
| Product |
Enzymatic Method |
Template |
Reference |
|
| Amine Labeling |
| Aminoallyl-dUTP |
Nick Translation |
Plasmid DNA |
Basu et al. (2014) |
|
| Hapten Labeling |
Biotin Labeling PCR Core Kit
(Biotin-16-dUTP – based) |
PCR |
Genomic DNA |
Garcia et al. (2015) |
| PCR fragment |
Melnikova et al. (2014) |
| Digoxigenin-11-dUTP |
PCR |
chromosomal DNA fragments |
Mattern et al. (2015) |
| Genomic DNA |
Tortajada-Genaro et al. (2016) |
|
| Fluorescent Labeling |
| Aminoallyl-dUTP-ATTO425 |
Nick Translation |
BAC clone |
Basinko et al. (2012) |
| Aminoallyl-dUTP-XX-ATTO488 |
Nick Translation |
BAC clone & genomic DNA |
Sanei et al. (2011) |
| PCR |
chromosomal DNA fragments |
Romanenko et al. (2015) |
| Whole genome amplified mitochondrial & plasmid DNA |
Seok et al. (2015) |
| Whole genome amplified BAC clone |
Boroviak et al. (2016) |
| Aminoallyl-dUTP-XX-ATTO532 |
PCR |
single-sorted chromosomal DNA fragments |
Potapova et al. (2011) |
| Aminoallyl-dUTP-ATTO550 |
PCR |
single-sorted chromosomal DNA fragments |
Potapova et al. (2011) |
| Aminoallyl-dUTP-Cy3 |
Nick Translation |
BAC clone and plasmid DNA |
Romanenko et al. (2015) |
| PCR |
chromosomal DNA fragments |
Vasconcelos et al. (2016) |
| Whole genome amplified BAC clone |
Boroviak et al. (2016) |
| Whole genome amplified mitochondrial & plasmid DNA |
Seok et al. (2015) |
| Aminoallyl-dUTP-TexasRed |
PCR |
Whole genome amplified mitochondrial & plasmid DNA |
Seok et al. (2015) |
| Whole genome amplified BAC clone |
Boroviak et al. (2016) |
| Aminoallyl-dUTP-Cy5 |
PCR |
chromosomal DNA fragments |
Romanenko et al. (2015) |
| Whole genome amplified mitochondrial & plasmid DNA |
Seok et al. (2015) |
| Whole genome amplified BAC clone |
Boroviak et al. (2016) |
Selected References:
Basinko et al. (2012) Clinical and molecular cytogenetic studies in ring chromosome 5: Report of a child with congenital abnormalities. European Journal of Medical Genetics 55:112e.
Basu et al. (2014) Using Amino-Labeled Nucleotide Probes for Simultaneous Single Molecule RNA-DNA FISH. PLOS ONE 9 (9):e107425.
Boroviak et al. (2016) Chromosome Engineering in Zygotes with CRISPR/Cas9. Genesis 54:78
Garcia et al. (2015) Reassignment of Drosophila willistoni genome scaffolds to chromosome II arms. Genes Genomes Genetics 5:2559.
Mattern et al. (2015) Identification of the antiphagocytic trypacidin gene cluster in the human-pathogenic fungus Aspergillus fumigatus. Appl. Microbiol. Biotechnol. 99:10151.
Melnikova et al. (2014) Retrotransposon-Based Molecular Markers for Analysis of Genetic Diversity within the Genus Linum. BioMed Research International 2014:1.
Morrison et al. (2003) Labeling Fluorescence In Situ Hybridization Probes for Genomic Targets. Methods in Molecular Biology 204:21.
Potapova et al. (2011) Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. Biotechniques 59 (6):335.
Romanenko et al. (2015) A First Generation Comparative Chromosome Map between Guinea Pig (Cavia porcellus) and Humans. PLOS ONE 10 (5):e0127937.
Sanei et al. (2011) Loss of centromeric histone H3 (CENH3) from centromeres precedes uniparental chromosome elimination in interspecific barley hybrids. PNAS 108 (33):E498.
Seok et al. (2015) Frequent somatic transfer of mitochondrial DNA into the nuclear genome of human cancer cells. Genome Research 25:814.
Tortajada-Genaro et al. (2016) Genotyping of single nucleotide polymorphisms related to attention-deficit hyperactivity disorder. Anal. Bioanal. Chem. 408:2339.
Vasconcelos et al. (2016) Intra- and interchromosomal rearrangements between cowpea [Vigna unguiculata (L.) Walp.] and common bean (Phaseolus vulgaris L.) revealed by BAC-FISH. Genome Research 23:253.
Wiegant et al. (1997) Probe Labeling and Fluorescence In Situ Hybridization. In: Current Protocols in Cytometry 8.3.1, John Wiley & Sons Inc.
|



 Jena Bioscience GmbH
Jena Bioscience GmbH 